
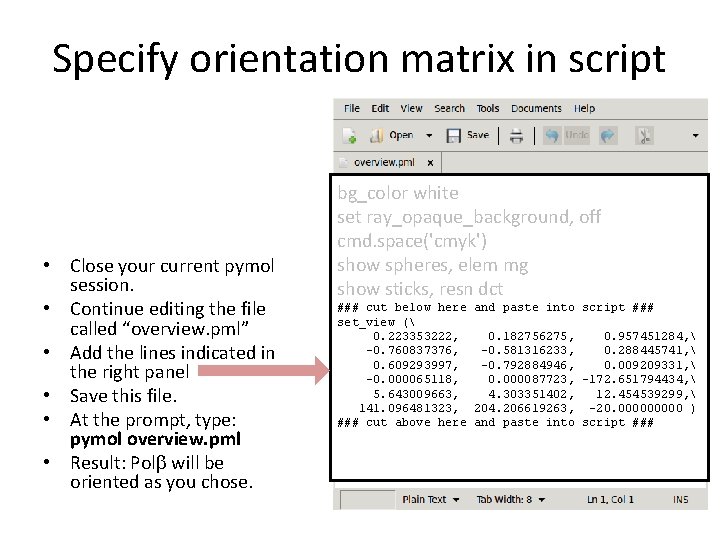
The PDB at and trying out its search function. Protein Data Bank, but you are better off just visiting The structure of "1xuu" or you can search the PDB (Protein Data Bank: Alternatively you can type "fetch 1xuu" in the command bar to download and display The PDB code for the structure you wish to download from the RCSB If you know the PDB ID of the structure you wish to look at, then theĮasiest way to load it into PyMOL is via the Plugin menu. I prefer a white background, especially for making figures for presentations so change this
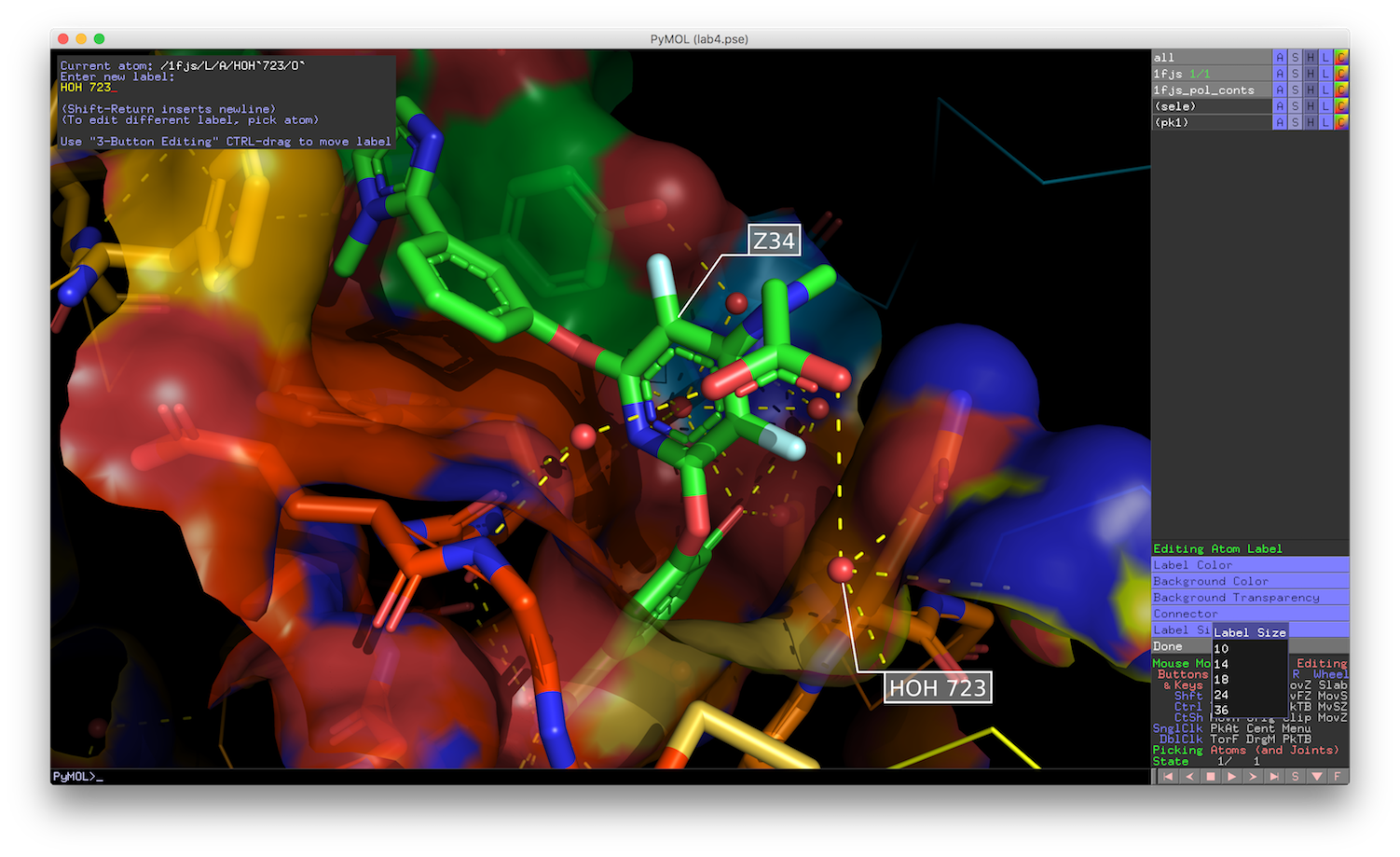
This (all the figures on this page link to larger images): May come talk to me to use one of our lab computers for the assignment.Ī good introduction to PyMOL can be found atĪfter installing PyMOL, start it up and you should see a window that looks like On his or her computer, may use a laptop in the Bracken Library elab or
#Pymol tutorial scripts mac osx#
Introduction to PyMOL PyMOL is a molecular graphics program thatĬan be freely downloaded and installed on Windows or Linux PCsĪs well as Apple computers running MAC OSX and other UNIX-based systems. This site will look much better in a browser that supports


 0 kommentar(er)
0 kommentar(er)
